Distinct baseline immune characteristics associated with responses to conjugated and unconjugated pneumococcal ... - Nature.com
Abstract
Pneumococcal infections cause serious illness and death among older adults. The capsular polysaccharide vaccine PPSV23 and conjugated alternative PCV13 can prevent these infections; yet, underlying immunological responses and baseline predictors remain unknown. We vaccinated 39 older adults (>60 years) with PPSV23 or PCV13 and observed comparable antibody responses (day 28) and plasmablast transcriptional responses (day 10); however, the baseline predictors were distinct. Analyses of baseline flow cytometry and bulk and single-cell RNA-sequencing data revealed a baseline phenotype specifically associated with weaker PCV13 responses, which was characterized by increased expression of cytotoxicity-associated genes, increased frequencies of CD16+ natural killer cells and interleukin-17-producing helper T cells and a decreased frequency of type 1 helper T cells. Men displayed this phenotype more robustly and mounted weaker PCV13 responses than women. Baseline expression levels of a distinct gene set predicted PPSV23 responses. This pneumococcal precision vaccinology study in older adults uncovered distinct baseline predictors that might transform vaccination strategies and initiate novel interventions.
Similar content being viewed by others
Baseline innate and T cell populations are correlates of protection against symptomatic influenza virus infection independent of serology
Distinct immunological and molecular signatures underpinning influenza vaccine responsiveness in the elderly

Response to the conjugate pneumococcal vaccine (PCV13) in patients with chronic lymphocytic leukemia (CLL)
Main
Streptococcus pneumoniae (pneumococcus) infections lead to life-threatening diseases and are responsible for thousands of hospitalizations and deaths annually in the United States1. Infants and older adults have the highest rates of invasive pneumococcal disease, with an ~10% mortality rate particularly affecting older adults according to reports from the Centers for Disease Control and Prevention reports. Two types of pneumococcal vaccines were approved by the Food and Drug Administration (FDA) for use in this population: the capsular polysaccharide vaccine Pneumovax® (PPSV23) and protein-polysaccharide conjugate vaccines (for example, Prevnar® (PCV13)). PPSV23 contains capsular polysaccharide antigens for 23 serotypes that elicit antibody responses in the absence of CD4+ T cell collaboration2. Responses to unconjugated polysaccharides are predominantly mediated by the activation of marginal zone (MZ) B cells, which possess hypermutated immunoglobulin genes and express high levels of complement receptor 2 (CD21)3,4. MZ B cells are not fully mature in newborns and children, resulting in limited success of unconjugated polysaccharide vaccines3,5,6. Furthermore, older individuals show a marked decrease in frequencies of total memory and MZ B cells, which might contribute to the age-related decline of responses7. After activation, MZ B cells terminally differentiate into IgM-, IgG- and IgA-secreting plasma cells8. Although MZ B cell activation and IgM production are considered to be T cell independent, IgG and IgA class switching might rely on the presence of non-cognate T cells that secrete IL-21 and/or trigger the CD40/CD40L signaling axis8. Conjugated vaccines have been developed (for example, PCV13) to enhance immunogenicity. PCV13 is composed of polysaccharides for the most prevalent 13 serotypes conjugated to a nontoxic variant of diphtheria toxin CRM197. PCV13 induces T cell-dependent activation and expansion of B2 follicular cells, promoting the generation of memory B cells and differentiation of long-lived plasma cells9,10,11.
Conjugated pneumococcal vaccines are effective among children and young adults1 and in preventing invasive disease among older adults12, yet their efficacy declines with age13,14,15. PCV13 is 90% effective in infants and children, which decreases to 72.8% among older adults. Similarly, the efficacy of PPSV23 is 54% for adults16. In 2021, other conjugated vaccines were approved, including PCV15 and PCV20, to extend the range of serotypes covered and to simplify pneumococcal immunizations17. In the United States, the current recommendation for older adults is to receive either PCV20 or PCV15 followed by PPSV23 1 year later.
Peripheral blood leukocyte gene expression studies revealed transcriptional differences in responses to PPSV23 and influenza vaccines among healthy adults18,19. However, these studies did not include conjugated pneumococcal vaccines and excluded older adults partially due to the complexity of evaluating their responses. To address these gaps, we recruited 39 healthy pneumococcal vaccine-naive adults ≥60 years old who received PCV13 or PPSV23. We collected blood from this cohort longitudinally and studied their baseline immune phenotypes and immune responses following vaccination by using multiple assays. Our systems vaccinology approach uncovered distinct baseline gene expression modules associated with PCV13 and PPSV23 responses, which can pave the way toward more precise administration of these vaccines.
Results
Serological responses of older adults to PCV13 and PPSV23
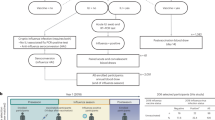
During the 2017–2018 seasons, 39 older adults (≥60 years of age with no history of pneumonia disease or vaccination) were recruited and randomized to receive either a dose of PCV13 (ten men and nine women) or PPSV23 (ten men and ten women). Vaccines were administered from May to early fall to avoid overlap with the influenza vaccine season. Donors in each arm of the study and the men and women in each arm were comparable in terms of age, body mass index (BMI), cytomegalovirus (CMV) seropositivity and frailty index (Fig. 1a). Donors were not on any confounding treatments, but some were on medications for common chronic diseases (Supplementary Table 1a,b).
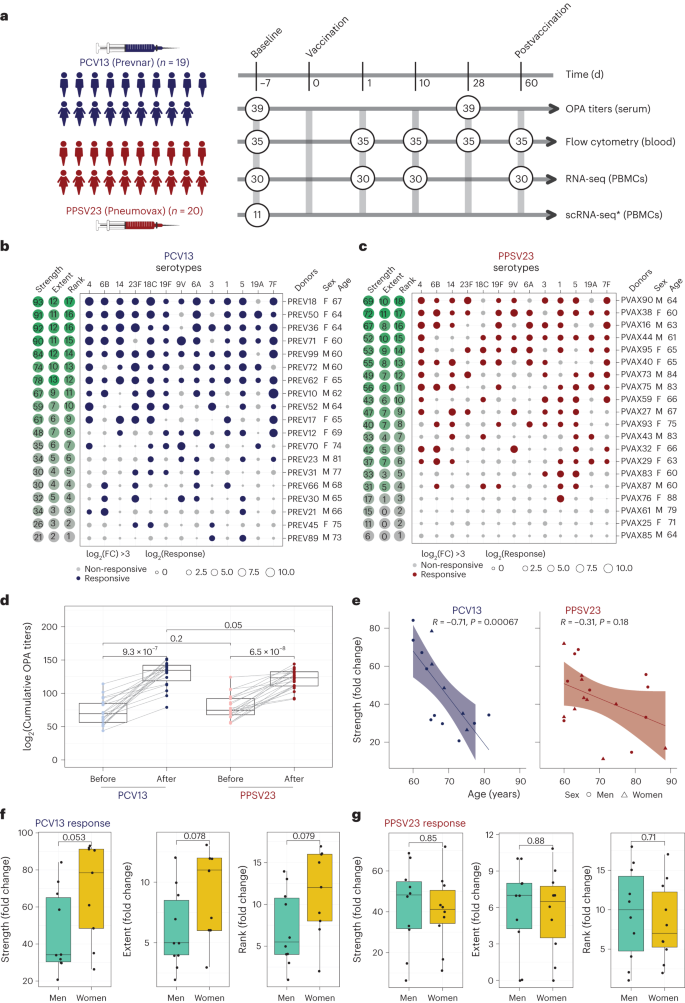
a, Schematic representation of the study design. Nine women and ten men received the PCV13 vaccine, and ten women and ten men received PPSV23. OPA titers for the 13 serotypes were assessed from serum samples obtained 7 d before vaccination (baseline) and 28 d after vaccination for both vaccines. Anticoagulated blood samples were used for flow cytometric analysis of whole-blood cell populations. PBMCs were isolated for bulk RNA-seq. Prevaccination PBMCs from four women and seven men who received PCV13 were isolated for scRNA-seq. The numbers in circles represent the total numbers of biologically independent samples processed for the indicated assays at the indicated times. Figure created with BioRender.com. The star indicates scRNA-seq data generated exclusively for the PCV13 baseline samples. b, Bubble plot of fold change (FC) in antibody titers for individual serotypes in response to PCV13 (n = 19); M, male; F, female. c, Bubble plot of fold change in antibody titers for individual serotypes in response to PPSV23 (n = 20). Dot size represents the fold change value, and color indicates a significant response (log2(fold change) is >3), with blue for PCV13 and red for PPSV23. Donors are ordered from top to bottom according to the vaccine response rank. On the left, the strength (log2(sum fold change)), extent of response (number of serotypes out of 13 to which an individual mounted a significant response) and rank are presented. d, Prevaccination and postvaccination cumulative OPA titers (expressed as sum log2) in response to PCV13 (n = 19) and PPSV23 (n = 20). e, Correlation analysis between the cumulative fold change (sum log2(fold change)) and age (in years). f,g, Sex-specific differences in strength, extent and rank in donors who received PCV13 (n = 19; f) and PPSV23 (n = 20; g). A Wilcoxon matched-pairs signed-rank test (two sided) was used in d to compare titers before and after vaccination with PCV13 or PPSV23. Box plots display the median and interquartile range (IQR; 25–75%), with whiskers representing the upper and lower quartiles ±1.5× IQR. A Wilcoxon rank-sum test (two sided) was used to compare strength, extent and rank between men and women vaccinated with PCV13 and PPSV23. The Pearson correlation metric was used to perform correlation analyses between strength and age (e), and P values were computed using two-sided t-tests; n represents the number of biological replicates.
Source data
Blood samples were collected 7 d before vaccination (baseline) and at 1, 10, 28 and 60 d after vaccination. Previously, we showed that transcriptional innate responses to PPSV23 peaked at day 1, whereas adaptive responses peaked at day 7 and remained high until day 14 in adults19. Hence, days 1 and 10 were chosen to capture the innate and adaptive responses while also ensuring participant safety and adherence to the institutional review board recommendations regarding the maximum volume of blood one can collect from older adults (550 ml over an 8-week period or 69 ml per week). We generated longitudinal flow cytometry and RNA-sequencing (RNA-seq) data to study immune responses to these two vaccines (Fig. 1a). Pneumococcal vaccine responses can be assessed by measuring antibody levels using enzyme-linked immunosorbent assays (ELISAs) or antibody function using opsonophagocytic activity (OPA). OPA measures the ability of antibodies to effectively opsonize bacteria and is ideal for quantifying responses because it mimics the in vivo mechanisms of antibody protection20 and is hence considered state of the art for FDA approvals21. Unlike children, many older adults already possess high ELISA antibody levels before vaccination, making functional OPA assays more attractive in this population20. Therefore, we used OPA to quantify antibody responses from serum (at baseline and 28 d after vaccination). Because OPA assays require a substantial volume of serum, we prioritized the 13 most prevalent serotypes: the 12 shared serotypes and serotype 6A from PCV13 (Supplementary Table 2)22. Quantifying the common serotypes enabled a side-by-side comparison of these two vaccines.
We developed three multivariate measures to quantify vaccine responses: (1) strength of the response (the sum of fold change values between baseline and day 28 for all serotypes), (2) extent (the number of serotypes (of 13) to which the donor elicits significant (that is, an OPA titer of ≥8) responses) and (3) rank (the rank of an individual's vaccine responsiveness in the cohort based on aggregate responses for all serotypes, where higher ranks represent stronger responses; Fig. 1b,c, Extended Data Fig. 1a,b and Supplementary Table 2d,h). To assess whether quantifying PPSV23 responses using 13 serotypes fully captures responsiveness to this vaccine, we reanalyzed OPA titers for all 23 serotypes from another study11, which revealed a strong and significant correlation between ranking using 13 serotypes and ranking using all serotypes (R = 0.91, P = 0.013; Extended Data Fig. 1g). Strength and extent of responsiveness were highly correlated with the rank, while capturing different aspects of vaccine responses (Extended Data Fig. 1c). BMI, frailty index and number of concomitant medications were not associated with responsiveness to either vaccine (Extended Data Fig. 1d and Supplementary Table 1a,b).
After vaccination, OPA titer levels significantly increased for both vaccines cumulatively (Fig. 1d) and for each serotype (Extended Data Fig. 1e). Stronger PCV13 responses were detected for serotype 4 and the PCV13-specific serotype 6A (Extended Data Fig. 1e). Although PPSV23 does not contain serotype 6A, it induced increased 6A responses potentially due to cross-reactivity with serotype 6B. Baseline OPA titers were comparable between the two cohorts (P = 0.2); however, individuals vaccinated with PCV13 had stronger responses (Fig. 1d; P = 0.05) even after excluding serotype 6A (P = 0.089; Extended Data Fig. 1f). Level of responsiveness varied from one individual to another. In the group treated with PCV13, top responder PREV18 (female, 67 years old) responded significantly to all serotypes except for 19A, whereas the bottom responder PREV89 (male, 73 years old) elicited significant responses to only two serotypes (Fig. 1b). In PPSV23, top responder PVAX90 (male, 64 years old) responded to ten serotypes, whereas the bottom responder PVAX25 (female, 71 years old) did not respond significantly to any of the tested serotypes (Fig. 1c). Our ranking strategy captured this heterogeneity at the donor level. There was a significant inverse correlation between baseline OPA titer levels and fold change in titers (Extended Data Fig. 2a–c), in alignment with previous studies23. There was a significant negative correlation between cumulative OPA titers at baseline and vaccine responsiveness rank (Extended Data Fig. 2d). Given this association, we calculated the maximum residual after baseline adjustment (maxRBA) values (Extended Data Fig. 1a,b), which correct for baseline titer levels by modeling fold increase in titer levels as an exponential function of the baseline titer levels. maxRBA and rank correlated significantly for both vaccines (Extended Data Fig. 1c).
Donor age was negatively associated with both strength (Fig. 1e; R = −0.71 and P = 0.00067 for PCV13, R = −0.31 and P = 0.18 for PPSV23) and rank (Extended Data Fig. 2d). Although male and female donors were comparable in terms of age, frailty and BMI (Supplementary Table 1b), women mounted stronger responses to the PCV13 vaccine (P = 0.053 for strength, P = 0.079 for rank; Fig. 1f), which was not observed for PPSV23 (Fig. 1g). Four of five top responders were women and four of five bottom responders were men in the group treated with PCV13. Overall, these data showed that both pneumococcal vaccines induced strong antibody responses; yet, at the individual level, the degree of responsiveness was variable. For PCV13, donor age significantly contributed to this variation, and there was a trend for stronger responsiveness for women. Interestingly, neither age nor sex was significantly associated with PPSV23 responsiveness.
PCV13 and PPSV23 induce similar transcriptional responses
We generated longitudinal bulk RNA-seq data at baseline and days 1, 10 and 60 from peripheral blood mononuclear cells (PBMCs) from 31 donors (15 PCV13 and 16 PPSV23), including strong responders (SRs) and weak responders (WRs) for each vaccine while also balancing female and male samples. Samples from 30 donors that passed quality control were used in differential analyses. Gene expression profiles after vaccination were compared to those at baseline (day −7; Supplementary Table 3). There were no significant transcriptional changes for days 1 or 60. However, at day 10, 50 and 41 genes were differentially expressed compared to baseline (false discovery rate (FDR) < 0.05), respectively, for PCV13 and PPSV23 (Extended Data Fig. 3a). Upregulated genes after vaccination included immunoglobulin heavy chain genes IGHG2 and IGHA2 and plasmablast-associated genes JCHAIN and MZB1 (Fig. 2a). The activity of these genes peaked at day 10 and returned to baseline levels by day 60 (Fig. 2b). A cumulative plasmablast activity score using plasma cell-specific genes19 (n = 33 genes) confirmed a significant and comparable increase in the expression of plasmablast-associated genes at day 10 for both vaccines that returned to baseline levels at day 60 (Fig. 2b). IGHG2, encoding the IgG2 isotype, was among the most upregulated genes for both vaccines (Fig. 2c). IGHA2, encoding the IgA2 isotype, was also upregulated for both vaccines (Fig. 2c and Extended Data Fig. 3b), in alignment with the serology data24.
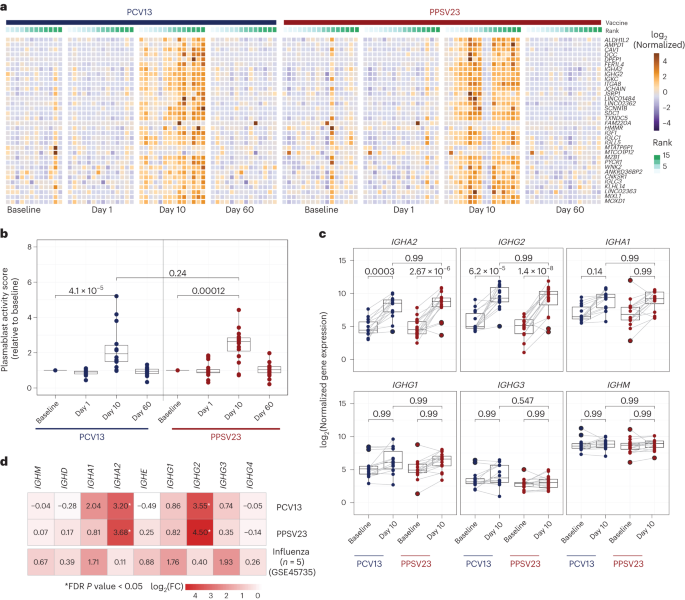
a, Heat map of differentially expressed genes between day 10 and baseline assessed using normalized gene expression values. b, Box plot of plasmablast activity scores at baseline and days 1, 10 and 60 in response to PCV13 (n = 14) and PPSV23 (n = 16), calculated using a published gene set (M4.11) and scaled with reference to baseline. c, Box plots showing normalized expression of genes encoding the constant region of the immunoglobulin heavy chain structure in response to PCV13 (n = 14) and PPSV23 (n = 16). d, Heat map showing differential expression of genes encoding the constant region of the immunoglobulin heavy chain structure at day 10 in response to PCV13 (n = 14) and PPSV23 (n = 16) and at day 7 in response to Fluzone (GSE45735 (influenza vaccine); n = 5). Genes with a >1.5-fold difference and FDR-corrected P value of <0.05 are marked with an asterisk (*). A Wilcoxon rank-sum test (two sided) was used to compare plasma cell activity scores between baseline and days 1, 10 and 60 (b). Box plots display the median and IQR (25–75%), with whiskers representing the upper and lower quartiles ±1.5× IQR. A Wilcoxon matched-pairs signed-rank test (two sided) was used to compare the expression of immunoglobulin genes at baseline and day 10 for PCV13 and PPSV23. FDR-corrected P values are shown in c; n represents the number of biological replicates.
Source data
Pneumococcal vaccines upregulated distinct immunoglobulin genes compared to the influenza vaccine. Reanalysis of prior PBMC RNA-seq data on influenza vaccine responses in younger adults showed upregulated expression of IGHG1 and IGHG3 at day 7, whereas pneumococcal vaccines upregulated the expression of IGHA2 and IGHG2 (Fig. 2d and Extended Data Fig. 3c). Transcriptional activation of plasmablast-associated genes significantly correlated with PCV13 responsiveness (R = 0.57, P = 0.034), mainly driven by IGHG2 expression (Extended Data Fig. 3d,f), which was not the case for PPSV23 (Extended Data Fig. 3e,f). Together, transcriptional profiling before and after vaccination revealed that both pneumococcal vaccines induced significant and similar plasmablast responses 10 d after vaccination, including increased transcription of genes encoding IgG2 and IgA2 antibody isotypes, contrasting with those (IgG1, IgG3 and IgA1) induced by the influenza vaccine.
Baseline frequencies of TH1 and TH17 cells are linked to PCV13 responses
We performed a longitudinal flow cytometry analysis of different cell populations in freshly isolated PBMCs at baseline (day −7) and after vaccination at days 1, 10, 28 and 60 to study B cell (naive, memory and plasmablast cells), CD4+ T cell (naive, type 1 helper T (TH1), type 2 helper T (TH2), interleukin-17 (IL-17)-producing helper T (TH17), IL-10-producing helper T (TH10), follicular helper T (TFH), type 1 TFH (TFH1), type 2 TFH (TFH2) and IL-17-producing TFH (TFH17) cells) and dendritic cell (DC) subsets (type 2 classical DCs (cDC2s), type 1 cDCs (cDC1s) and plasmacytoid DCs (pDCs); Supplementary Table 4a and Extended Data Fig. 4a,b). Despite the robust plasmablast transcriptional signature, their absolute numbers were only marginally increased at day 10 for both vaccines (Fig. 3a, top). There were no significant changes in other B cell subsets after vaccination or between the two cohorts (Extended Data Fig. 4b). Among the studied T cell subsets, only ICOS+ TFH cell numbers significantly increased at day 10 for both vaccines (Fig. 3a, bottom, and Extended Data Fig. 4b). There were no significant changes in other T cell or DC subsets after vaccination (Extended Data Fig. 4b). Cell numbers were similar between the two cohorts for all cell types both at baseline and after vaccination (Extended Data Fig. 4b). Therefore, the most significant changes included an increase in plasmablasts and ICOS+ TFH cells at day 10, which was comparable between the two vaccines. However, unlike our previous observations for influenza vaccines25, changes in these cell types did not correlate with vaccine responsiveness (Extended Data Fig. 4c).
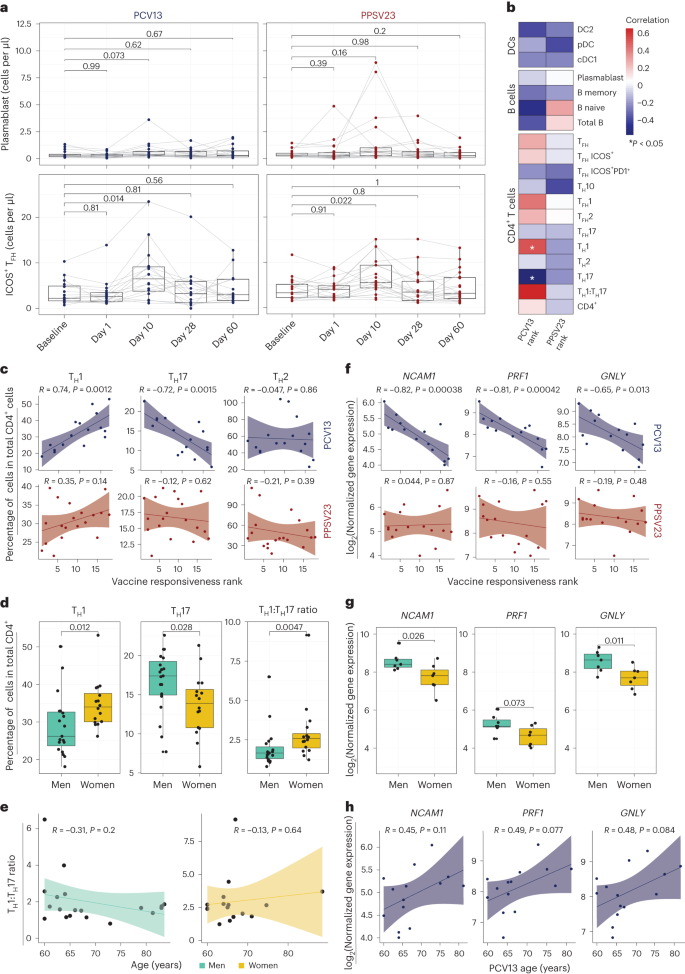
a, Longitudinal analysis of the absolute numbers of plasmablasts (cells per μl; top) and ICOS+ TFH cells (cells per μl; bottom) among the memory CD4+ T cell population in response to PCV13 (n = 16) and PPSV23 (n = 19). b, Correlation analysis between the absolute number of different cell types (DC, B cell and CD4+ T cell subsets) analyzed in whole blood and ranks. c, Correlation analysis between ranks and frequencies of TH1, TH17 and TH2 cells evaluated at baseline. d, Sex differences in the frequencies of TH1 and TH17 cells and TH1:TH17 cell ratio at baseline (n = 16 for PCV13; n = 19 for PPSV23). TH1 and TH17 cell frequencies were calculated relative to the total CD4+ T cell count. e, Association between TH1:TH17 cell ratio and age among men (green) and women (dark yellow) in response to PCV13 and PPSV23. f, Correlation analysis between baseline expression of cytotoxic genes (NCAM1, GNLY and PRF1) and PCV13 vaccine responsiveness rank (top; n = 14) and PPSV23 vaccine responsiveness rank (bottom; n = 16). g, Sex differences in the expression of NCAM1, PRF1 and GNLY at baseline (n = 14 for PCV13; n = 16 for PPSV23). h, Association between NCAM1, PRF1 and GNLY expression and age (n = 14 for PCV13; n = 16 for PPSV23). Box plots display the median and IQR (25–75%), with whiskers representing the upper and lower quartiles ±1.5× IQR. A Wilcoxon matched-pairs signed-rank test (two sided) was used to compare the absolute numbers of plasmablasts and ICOS+ TFH cells longitudinally (a). Correlation analyses were performed using the Pearson correlation metric (b, c, e, f and h), and P values were computed using two-sided t-tests; n represents the number of biological replicates.
Source data
To uncover baseline cellular predictors, we performed correlation analyses between the prevaccination absolute cell numbers for B cell, T cell and DC subsets and vaccine responsiveness (rank) for each vaccine. None of the profiled cell types were significantly associated with PPSV23 (Fig. 3b). By contrast, TH1 cell frequency (R = 0.74, P = 0.0012) was positively associated with PCV13 responses, while proinflammatory TH17 cell frequency (R = −0.72, P = 0.0015) was negatively associated with PCV13 responses (Fig. 3b,c; see Extended Data Fig. 5a,b for cell numbers). The ratio of TH1 cell frequency to TH17 cell frequency was the strongest predictor of PCV13 responses (R = 0.63, P = 0.0089) but not PPSV23 responses (Fig. 3b and Extended Data Fig. 5c). There was no significant difference in TFH frequency between SRs and WRs, although TFH1 cells showed a positive association with PCV13 responsiveness, and TFH17 cells showed a negative association with PCV13 responsiveness (Fig. 3b). We observed sex differences in TH1 and TH17 cell frequencies; women had higher TH1 (P = 0.012) and lower TH17 cell frequencies (P = 0.028) than men (Fig. 3d and Extended Data Fig. 5c). Women with higher TH1:TH17 cell ratios mounted stronger responses to PCV13 (Extended Data Fig. 5c). These two cell types and their ratios were not significantly associated with age (Fig. 3e). Together, these data established that individuals with higher frequencies of TH1 cells and lower frequencies of TH17 cells before vaccination mount stronger responses to the conjugated PCV13 vaccine.
Baseline CYTOX level is negatively associated with PCV13 responses
To identify baseline transcriptional signatures associated with vaccine responsiveness, we conducted weighted gene coexpression network analysis (WGCNA) on PBMC RNA-seq data, which yielded 33 gene modules. We associated these modules with vaccine responsiveness, sex, age and TH1:TH17 cell ratio. Of 33 modules, only 1 (midnight blue, 260 genes) was significantly (P = 0.02) and specifically associated with PCV13 responses (Extended Data Fig. 6a and Supplementary Table 5a). Genes in this module were enriched in the 'natural killer cell mediated cell cytotoxicity' KEGG pathway (P = 0.000003639) and included NCAM1, the marker gene for natural killer (NK) cells, and several transcripts (GNLY, PRF1 and GZMB) encoding cytotoxic functions26 (Supplementary Table 6a). This cytotoxicity-associated gene module is hereafter referred to as the CYTOX module. Baseline expression levels of CYTOX genes in PBMCs was negatively associated with responsiveness to PCV13 (for example, for NCAM1, R = −0.82 and P = 0.00038) but not responsiveness to PPSV23 (Fig. 3f). In addition, on average, PBMCs from men showed higher expression of CYTOX genes than PBMCs from women in both cohorts (Fig. 3g and Supplementary Table 6a). Expression levels of the top CYTOX genes increased with age, albeit not significantly (Fig. 3h); age-related upregulation of these genes was also observed in a larger cohort (n = 41 men and 34 women; P < 0.05 (ref. 27); Extended Data Fig. 6b). A different module (dark green, 134 genes) was specifically associated with reduced PPSV23 responses (P = 0.05; Extended Data Fig. 6a,c and Supplementary Table 6b), including genes associated with the cell cycle and transcriptional regulation (ANGEL2, MRE11, MTERF1, NUP107 and YTHDC2); however, they were not associated with any immune cell type or function. The top genes in this module showed no significant association with age and sex (Extended Data Fig. 6d,e). To summarize, WGCNA of the baseline PBMC RNA-seq data revealed two distinct gene sets that are significantly associated with PCV13 and PPSV23 responses.
The CYTOX signature stems from CD16+ NK cells
To identify cells bearing the CYTOX signature, we generated single-cell RNA-seq (scRNA-seq) data from prevaccination PBMCs of 11 PCV13 donors (6 SRs and 5 WRs), resulting in 52,702 cells after quality control (Fig. 4a and Supplementary Table 7). Clustering of PBMCs generated 24 clusters, which were annotated into monocytes (CD14 and CD16), DCs (monocyte-derived DCs, cDC1s and cDC2s), B cells (naive cells, memory cells, switched memory B cells, age-associated B cells and plasma cells), CD4+ T cells (naive, memory, regulatory T (Treg) and cytotoxic CD4+ T cells), CD8+ T cells (naive and memory cells (granzyme K+ T cells, effector memory T cells expressing CD45RA (TEMRA), mucosal-associated invariant T cells and γδ T cells)) and NK cells (CD56brightCD16− and CD56dimCD16+; Fig. 4a and Extended Data Fig. 7a,b). Among these, only two cell subsets were significantly different between SRs and WRs (Fig. 4b and Extended Data Fig. 8a), where SRs had more naive CD8+ T cells (P = 0.03) and fewer CD16+ NK cells (P = 0.03; Fig. 4b and Extended Data Fig. 8a).
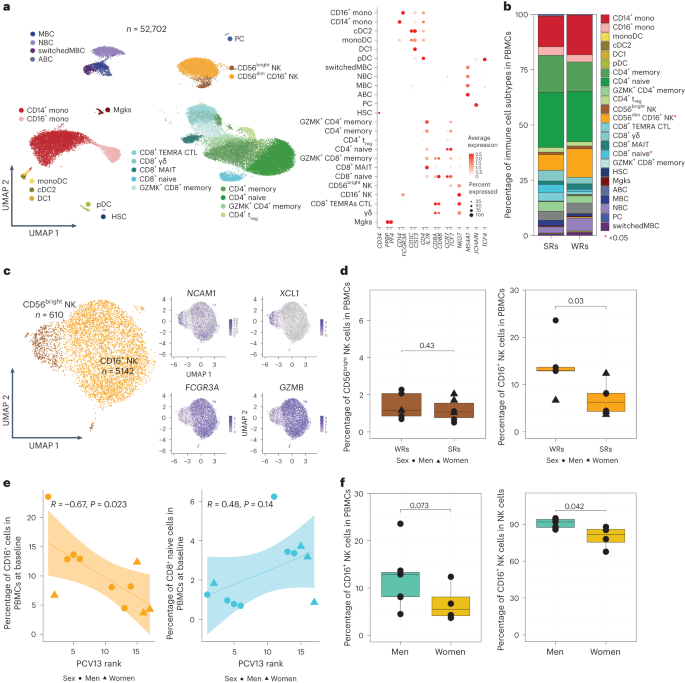
a, Uniform manifold approximation and projection (UMAP) of PBMCs from 11 PCV13 donors (six SRs and five WRs) showing 24 clusters from 52,702 cells colored by immune cell type. Immune subsets were identified in a supervised manner. Lineage markers are shown in the dot plot; MBC, memory B cell; NBC, naive B cell; ABC, age-associated B cell; mono, monocyte; monoDC, monocyte-derived DC; HSC, hematopoietic stem cell; Mgk, megakaryocyte; MAIT, mucosal-associated invariant T cell; GZMK, granzyme K; PC, plasma cells; CTL, cytotoxic T cell. b, Stacked bar plot of immune cell frequencies in SRs and WRs. The cell types with significant differences in their frequencies between SRs and WRs are marked with a red asterisk (*; P < 0.05). c, UMAP of NK cell subsets with feature plots showing the expression NCAM1, XCL1, FCGR3A and GZMB in blue, highlighting the two NK populations: CD56dimCD16+ NK cells and CD56bright NK cells. d, Box plots of CD16+ NK cell and CD56bright NK cell frequencies in SRs (n = 6) and WRs (n = 5). e, Correlation analysis between PCV13 rank and prevaccination frequencies of CD16+ NK and CD8+ naive T cells (n = 11). f, Sex differences in the prevaccination percentages of CD16+ NK cells in total PBMCs and in total NK cells (n = 11). Box plots display the median and IQR (25–75%), with whiskers representing the upper and lower quartiles ±1.5× IQR. A Wilcoxon rank-sum test (two sided) was used to compare cell percentages between SRs and WRs (b and d) and CD16+ NK cell percentages between men and women (f). Correlations were computed using the Pearson correlation metric (e), and P values were computed using two-sided t-tests; n represents the number of biological replicates.
Source data
Blood NK cells include immature CD56brightCD16− NK cells that produce the chemokine XCL1 and the mature, highly cytotoxic CD56dimCD16+ cells, both of which were captured in the scRNA-seq data (Fig. 4c). However, only the CD16+ NK cells were significantly associated with vaccine responsiveness, where WRs had higher frequencies of these cells (Fig. 4d,e and Extended Data Fig. 8b). In line with their stronger PCV13 responses (Fig. 1f), women had lower CD16+ NK cell frequencies than men (P = 0.042; Fig. 4f). We calculated CYTOX scores at single-cell resolution using the average expression of CYTOX genes. The CYTOX score was highest in the NK cell populations, followed by the cytotoxic CD8+ T cell populations (Extended Data Fig. 9a). There was no significant difference between the CYTOX scores of SRs and WRs neither for the memory CD4+ T cell clusters (Extended Data Fig. 9b–e) nor for other cell types, including TEMRAs that have high CYTOX scores (Extended Data Fig. 9e). Together, these data establish that CD16+ NK cells comprise the population of cells that bears the CYTOX signature, where the increased frequency of CD16+ NK cells is associated with reduced PCV13 responses.
Differential gene expression analyses between CD16+ NK cells of SRs and WRs revealed 38 differentially expressed genes (FDR of 5%; Fig. 5a and Supplementary Table 8). CD16+ NK cells from WRs showed higher expression of cytotoxic genes (GNLY and GZMH) and IFNG, indicating a more 'cytotoxic' and 'activated' phenotype. Furthermore, WR CD16+ NK cells expressed higher levels of the inhibitory receptor gene KLRC2 (NKG2C), suggesting a more advanced maturation stage28. CD16+ NK cells from WRs also overexpressed amphiregulin (AREG), a ligand for the epidermal growth factor receptor that is upregulated after inflammation and mitochondrial stress in immune cells29. Although CMV can modulate NK cell phenotypes30, CMV positivity (10 of 39 donors) was not associated with responsiveness to either vaccine (Fig. 5b). These data suggest that CD16+ NK cells from PCV13 WRs had a more cytotoxic and activated transcriptional phenotype than the same cells from SRs.
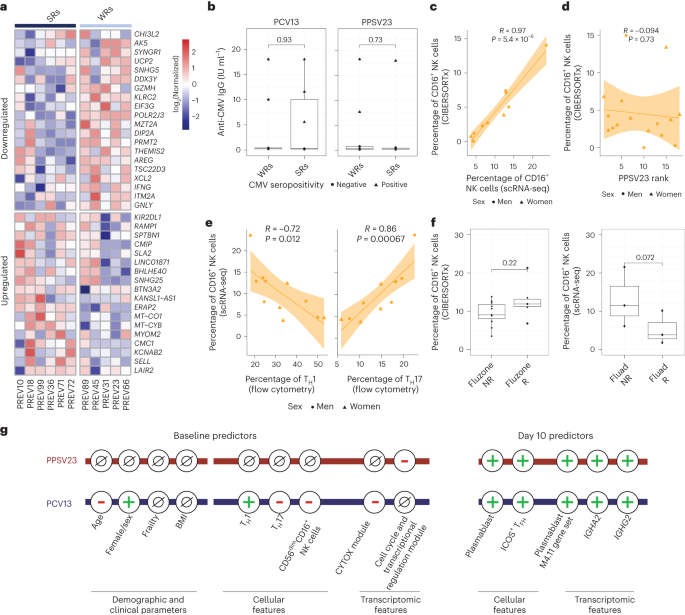
a, Heat map of the differentially expressed genes in CD16+ NK cells of six PCV13 SRs and five WRs at baseline, as assessed using normalized expression values from the scRNA-seq pseudobulk analysis. b, Box plots comparing anti-CMV IgG titers between SRs and WRs for PCV13 (left) and PPSV23 (right). c, Correlation analysis of prevaccination CD16+ NK cell percentages estimated by scRNA-seq and CIBERSORTx (n = 11). d, Correlation analysis of CIBERSORTx-based estimates of CD16+ NK cells and PPSV23 rank (n = 16) at baseline. e, Correlation analysis of CD16+ NK cell percentages determined by scRNA-seq and TH1 and TH17 cell percentages determined using flow cytometry at baseline. f, Box plots of prevaccination CD16+ NK cell percentages in Fluad responders (R; n = 3) and non-responders (NR; n = 3) and Fluzone trivalent inactivated influenza vaccine responders (n = 5) and non-responders (n = 11). g, Summary schema showing the demographic, clinical, cellular and transcriptomic parameters associated with PCV13 and PPSV23 vaccine responsiveness at baseline and day 10. Box plots display the median and IQR (25–75%), with whiskers representing the upper and lower quartiles ±1.5× IQR. A Wilcoxon rank-sum test (two sided) was used to compare the mean anti-CMV IgG titers between SRs and WRs of PCV13 and PPSV13 donors (b) and prevaccination CD16+ NK cell percentages in Fluad responders and non-responders and Fluzone responders and non-responders (f). Correlation analyses were computed using the Pearson correlation metric (c, d and e), and P values were computed using two-sided t-tests; n represents the number of biological replicates.
Source data
PPSV23 responses are independent of CD16+ NK cell frequency
To establish whether CD16+ NK cells are also associated with PPSV23 responsiveness, we used CIBERSORTx and inferred cell-type frequency from bulk data by using cell-type-specific expression from scRNA-seq data (Fig. 4a). First, we confirmed that inferred CD16+ NK cell frequencies match scRNA-seq-based frequencies using 11 donors for whom we have both scRNA-seq and bulk PBMC RNA-seq data (R = 0.97, P = 5.4 × 10–6; Fig. 5c and Extended Data Fig. 10a). In the PCV13 cohort, inferred CD16+ NK cell frequency significantly correlated with responsiveness (R = −0.69, P = 0.0067), reinforcing our observation. By contrast, the CD16+ NK cell frequency was not associated with PPSV23 responsiveness (R = −0.094, P = 0.73; Fig. 5d and Extended Data Fig. 10b). Together with the lack of association between CYTOX genes and PPSV23 responsiveness (Fig. 3e), these analyses established that CD16+ NK cells are important only for responses to the conjugated PCV13 vaccine. Notably, donors with higher CD16+ NK cell frequencies also had higher TH17 cell frequencies (R = 0.86, P = 0.00067) and lower TH1 cell frequencies (R = −0.72, P = 0.012; Fig. 5e), which was not observed for CD16− NK cells (Extended Data Fig. 10c).
To study the role of CD16+ NK cells in responses to other vaccines in older adults, we reanalyzed bulk PBMC RNA-seq data from the previously published Fluzone study (n = 16, 65+ years old)31. CIBERSORTx-inferred CD16+ NK cell frequencies did not show significant differences between responders and non-responders (P = 0.22; Fig. 5f). However, reanalyses of scRNA-seq data from individuals treated with the adjuvanted influenza vaccine Fluad (n = 6, 65 years old)32 showed that CD16+ NK cell frequencies were higher in non-responders than in responders (three responders and three non-responders; P = 0.072; Fig. 5f). Together, these...
Comments
Post a Comment